Research
The Wang lab integrates advanced sequencing and high-performance computational genomics to enable precision medicine in diverse healthcare settings and populations. Our main focus is development of technologies to support faster, more accurate, and resource-appropriate molecular diagnostics for cancer and microbial pathogens.
As a part of UNC RAISE, a primary goal is the development of sequencing and computational technologies to enable precision cancer diagnostics in low-resource settings
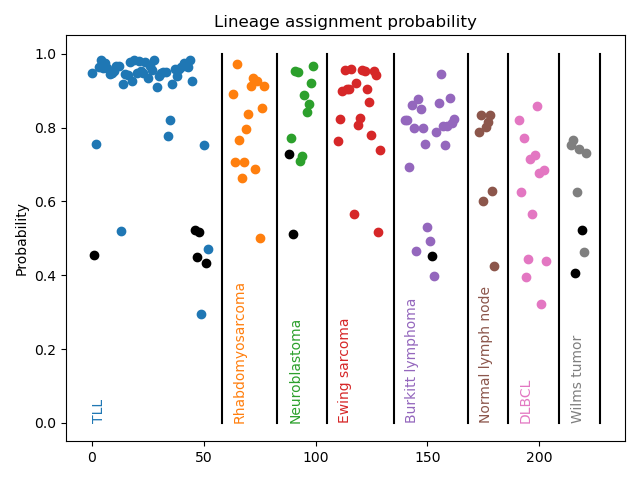
Pediatric cancer classification
Comprehensive clinical diagnosis of childhood cancers relies on multiple costly and time-consuming techniques such as immunohistochemistry (IHC), flow cytometry, cytogenetics, fluorescence in situ hybridization (FISH), targeted PCR panels, and microarrays. Nanopore RNA-sequencing represents a potential alternative to flow cytometry, IHC, and cytogenetic approaches in low-resource settings. Clinically-relevant characterization, including cell lineage and genomic subtype, are made from low-coverage transcriptome profiling via a machine learning classifier.
Metagenomics and genomic epidemiology
Single-molecule sequencing of metagenomes produces more specific taxonomic and genic characterization than standard NGS, but challenges remain due to relatively low throughput and high error rate, particularly from host-associated microbial communities. We have developed novel sequencing and informatics approaches to increase throughput and classification accuracy in both synthetic and human tissue-associated microbiota.
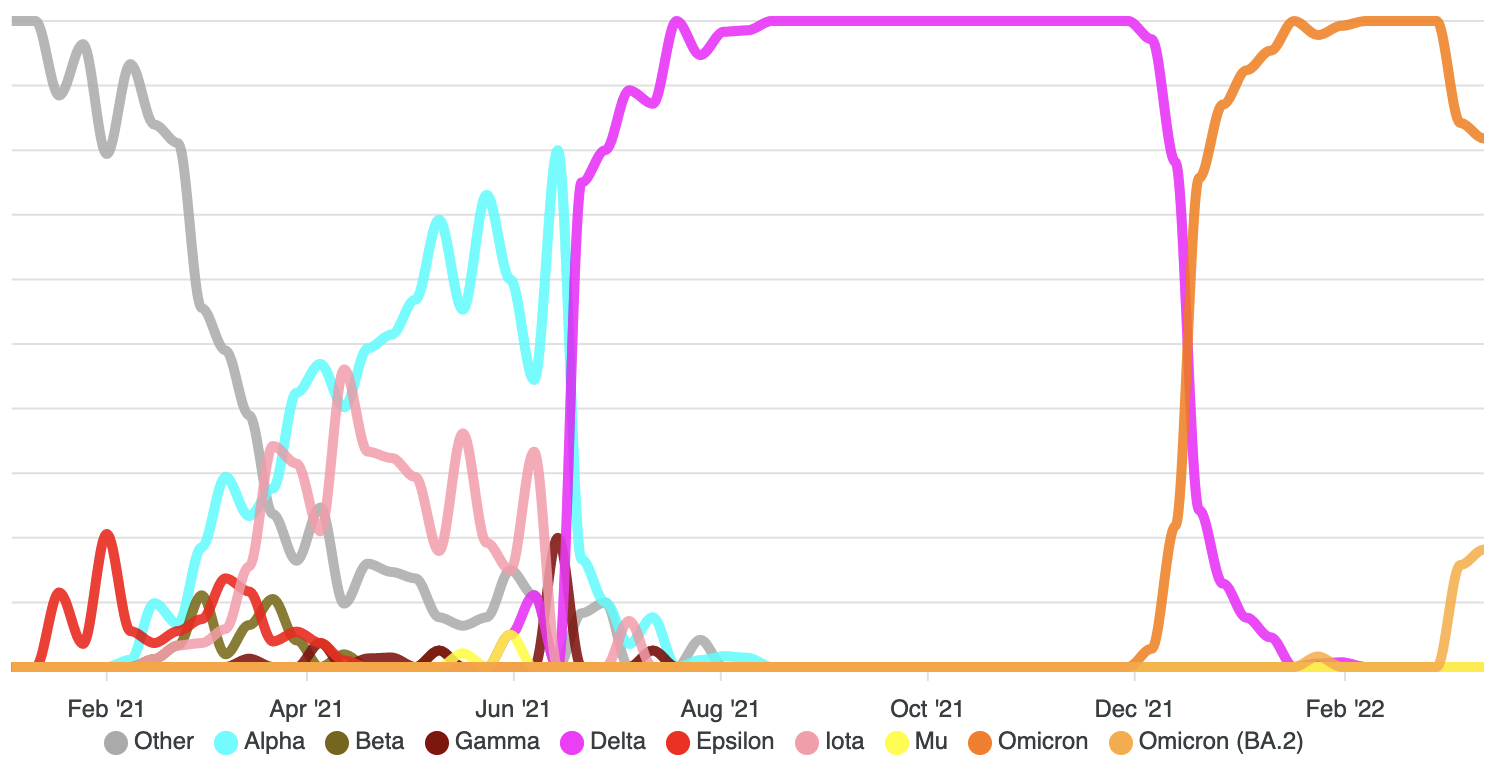
Genomic epidemiology of SARS-CoV-2 and anti-microbial resistance
Recent Publications
Pediatric cancer genomics and diagnostics
Microbial genomics and epidemiology
People

Associate Professor
Department of Pathology and Laboratory Medicine
Department of Genetics
UNC Lineberger Comprehensive Cancer Center
Program for Precision Medicine in Health Care

Ph.D. Student
Bioinformatics and Computational Biology

Research Associate

Ph.D. Student
Pathobiology and Translational Science

Pediatric Hematology-Oncology Fellow

Visiting Postdoctoral Scholar
Barretos Cancer Hospital

Undergraduate Researcher

Graduate Rotation Student
Biological & Biomedical Sciences Program

Research Specialist

Research Professional/Project Manager
Alumni and Friends
Kofi Opoku, M.D., M.P.H.
Research Associate
Bryce Menichella
Undergraduate Researcher
Ashley Merrills
Undergraduate Researcher
Liliana Bolaños Abarca
Undergraduate Researcher
Claire Worsham
Undergraduate Researcher
Ana Flávia Péres
Visiting Ph.D. Student
Cornelia Roata
Undergraduate Researcher

Prospective students & postdocs
Graduate students are admitted through the Biological & Biomedical Sciences Program, through which you may join any number of labs, including ours.
Prospective postdocs should email their CV to [ jeremy @ unc ⋅ edu ].