Research
The Wang lab integrates advanced sequencing and high-performance computational genomics to enable precision medicine in diverse healthcare settings and populations. Our main focus is development of technologies to support faster, more accurate, and resource-appropriate molecular diagnostics for cancer and microbial pathogens.
As a part of UNC RAISE, a primary goal is the development of sequencing and computational technologies to enable precision cancer diagnostics in low-resource settings
Pediatric cancer classification
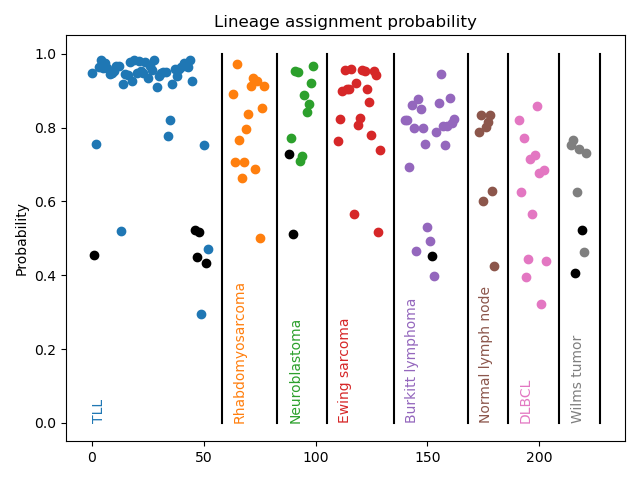
Comprehensive clinical diagnosis of childhood cancers relies on multiple costly and time-consuming techniques such as immunohistochemistry (IHC), flow cytometry, cytogenetics, fluorescence in situ hybridization (FISH), targeted PCR panels, and microarrays. Nanopore RNA-sequencing represents a potential alternative to flow cytometry, IHC, and cytogenetic approaches in low-resource settings. Clinically-relevant characterization, including cell lineage and genomic subtype, are made from low-coverage transcriptome profiling via a machine learning classifier.
Wang et al, 2022Metagenomics and genomic epidemiology
Single-molecule sequencing of metagenomes produces more specific taxonomic and genic characterization than standard NGS, but challenges remain due to relatively low throughput and high error rate, particularly from host-associated microbial communities. We have developed novel sequencing and informatics approaches to increase throughput and classification accuracy in both synthetic and human tissue-associated microbiota.
Wang et al, 2021Genomic epidemiology of SARS-CoV-2 and anti-microbial resistance
Wang et al. 2022. Tegha et al. 2020.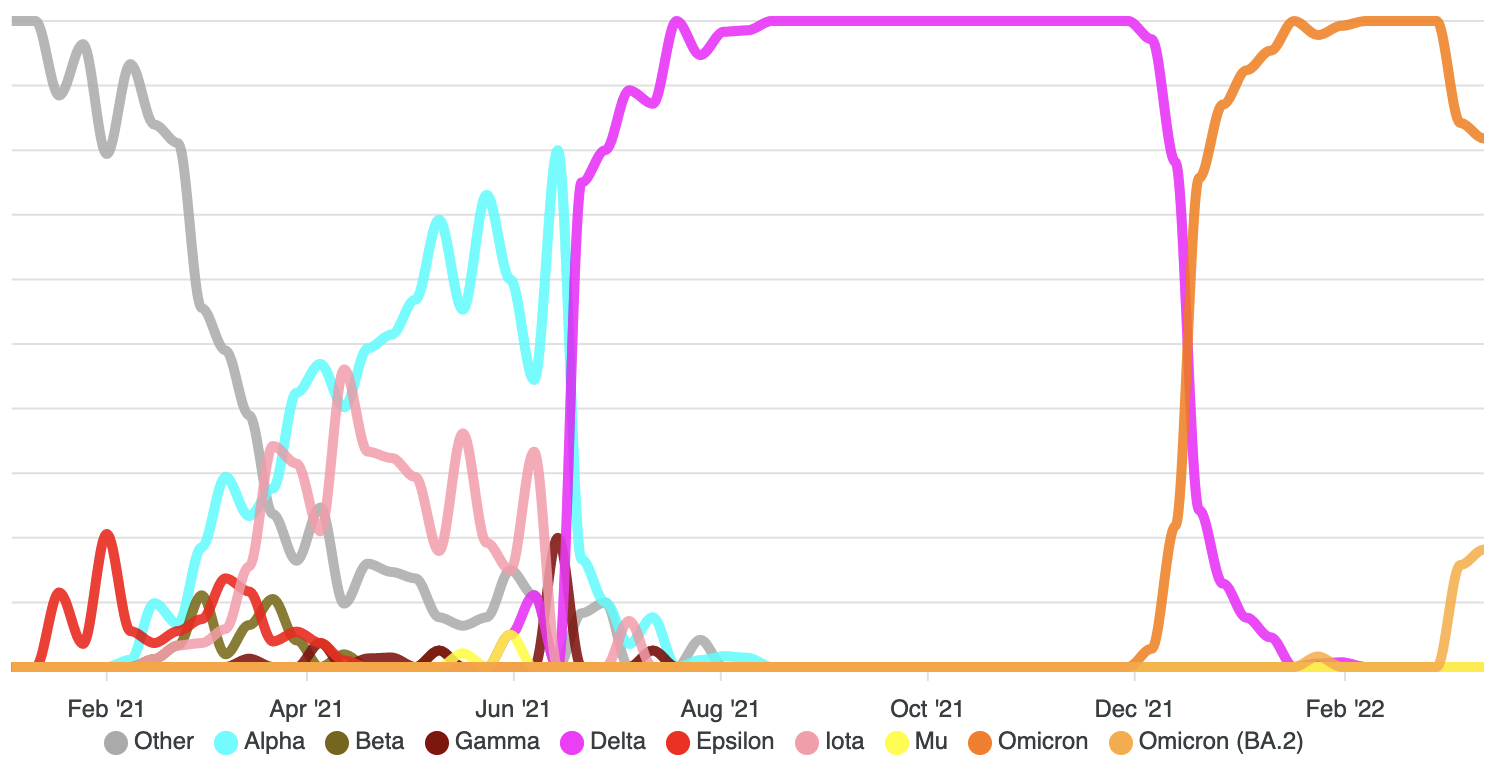
Recent Publications
Pediatric cancer genomics and diagnostics
Microbial genomics and epidemiology





